library(tidyverse)
band_members
band_instruments10. Joins
Video Tutorial
Joining two Tables together
Joins are powerful functions that allow you to connect two datasets together through matching values. They can be useful with spatial and non-spatial data alike.
Joins rely on “keys” that match records across different datasets. This can be something like a name or ID number.
As a recap of how joins work, we’re going to show a simple example of two different kinds of joins: ‘left’ joins and ‘inner’ joins. For this example, we will be using the band_members and band_instruments dataframes, which are simple, 3-row datasets that comes included in the dplyr package.
As you would expect, the first two arguments in a join function are the two tables you are trying to connect together. The third argument is the “join field”, which is the matching column in both datasets we will use to pair up rows.
Left Joins
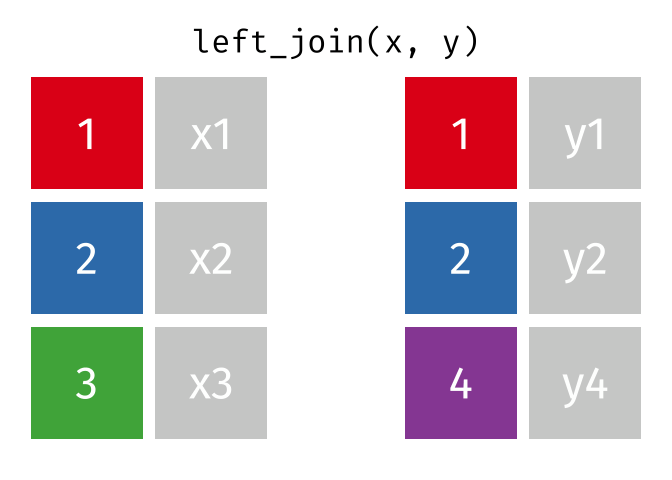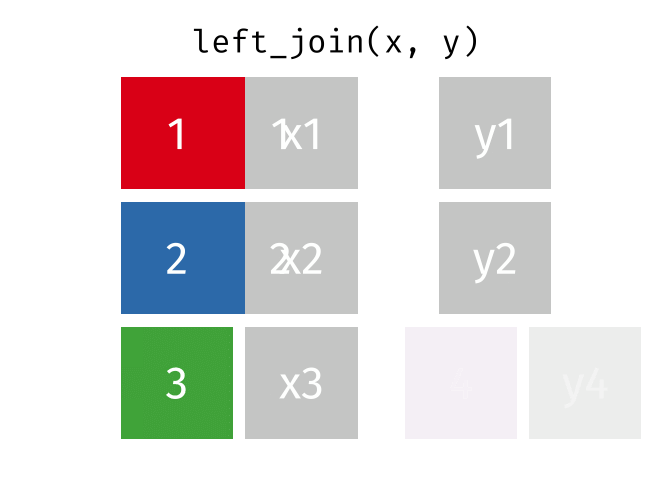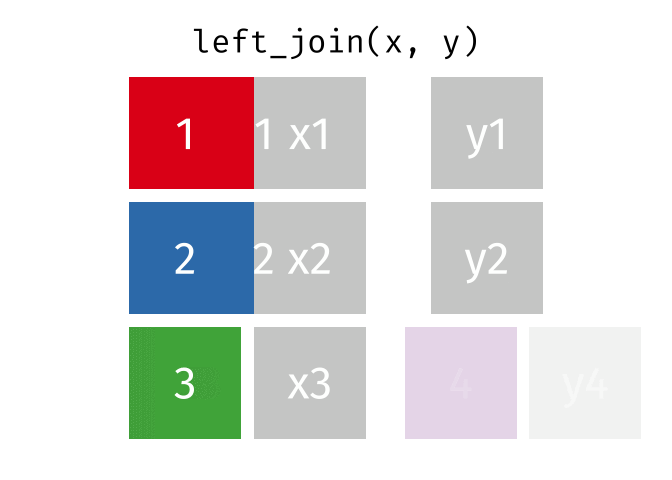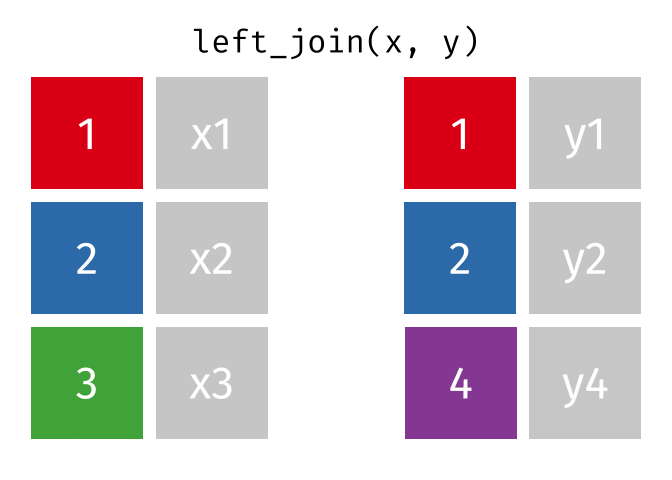
A left_join keeps all of the rows in the first table you specify, appending data from the second table through matching values in the specified “join field”. Let’s see how this kind of join looks with our example data:
band_members_and_instruments <-
left_join(band_members, band_instruments, by = "name")
# When the "join field" column names don't match, you can use:
# by = c("column1" = "column2")
band_members_and_instrumentsInner Joins
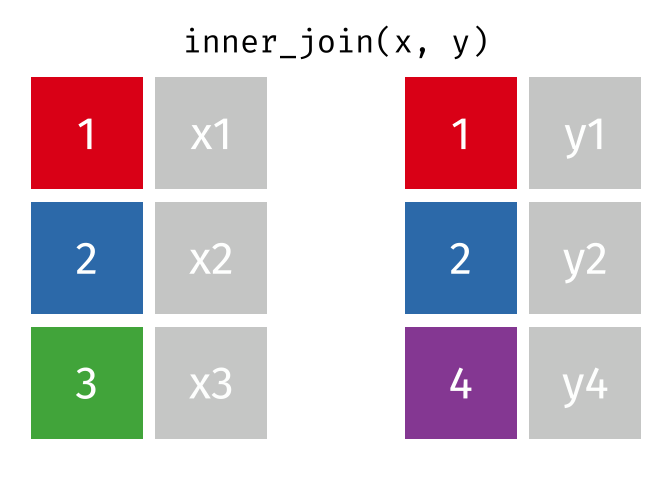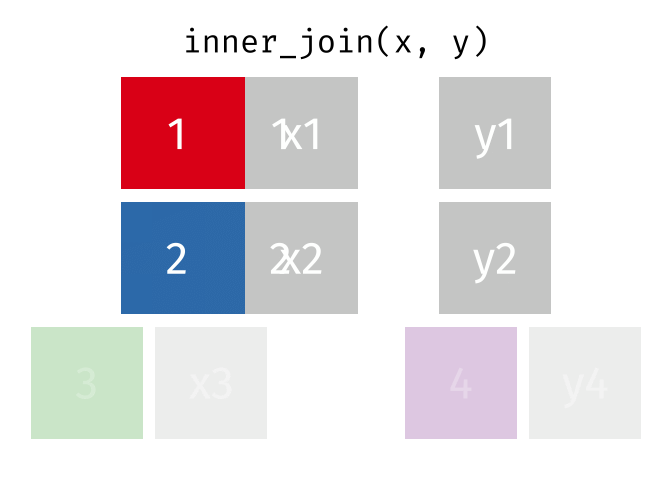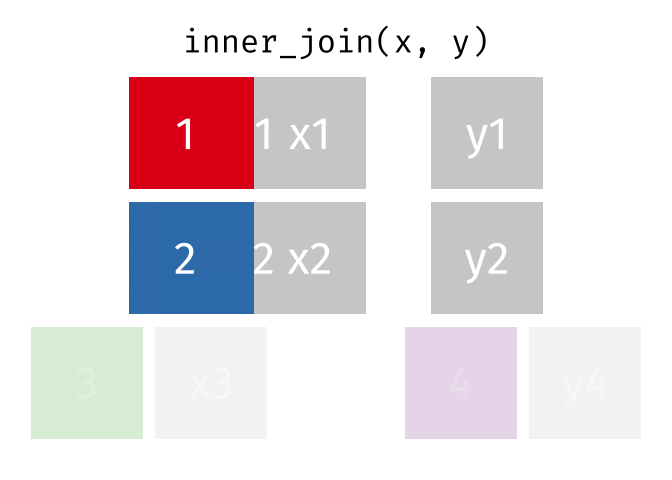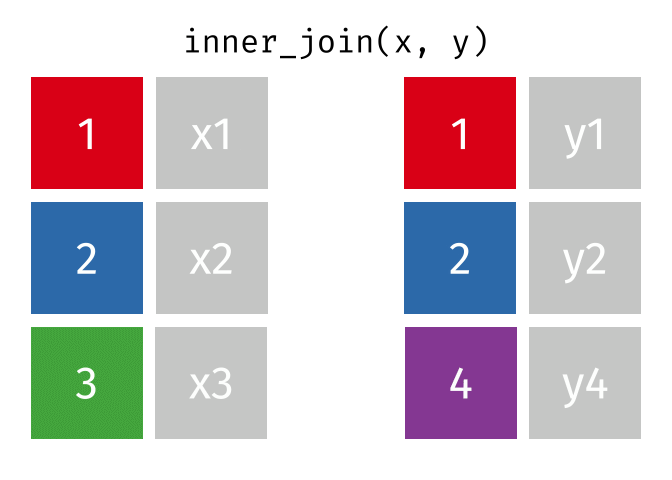
An `inner_join` keeps only the rows that have matching values between both tables in the specified “join field.” Any other rows are discarded. Let’s see how this kind of join looks with our example data:
band_members_with_instruments_only <-
inner_join(band_members, band_instruments, by = "name")
band_members_with_instruments_onlyFull Joins
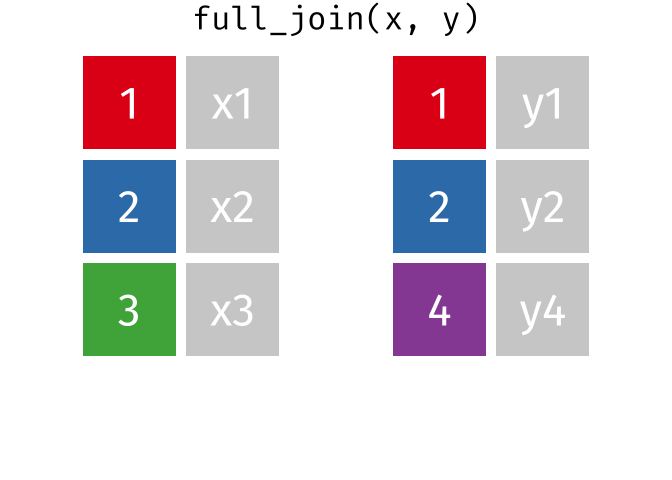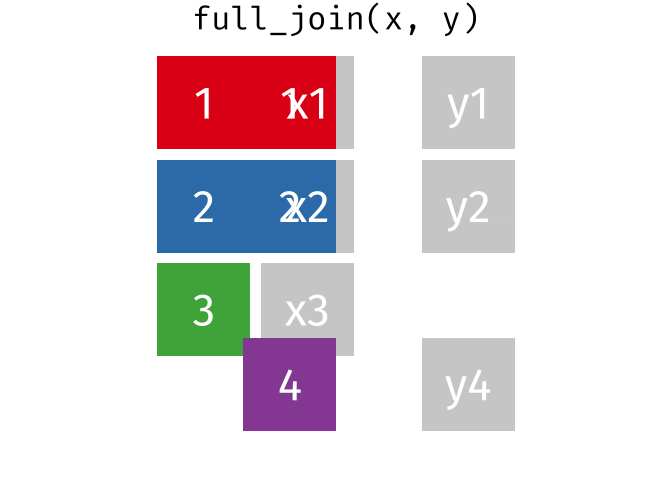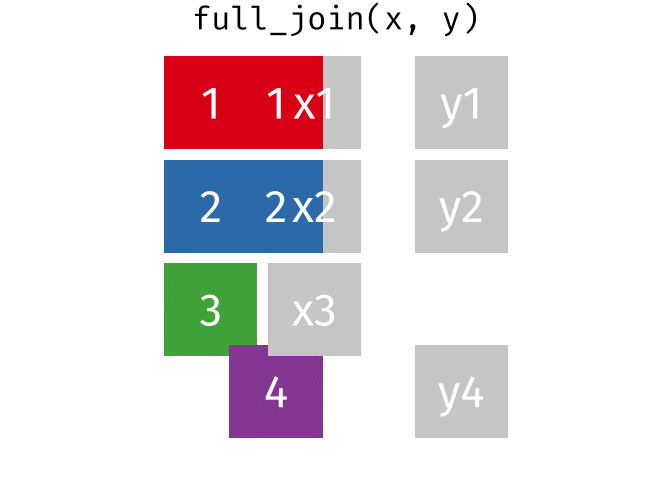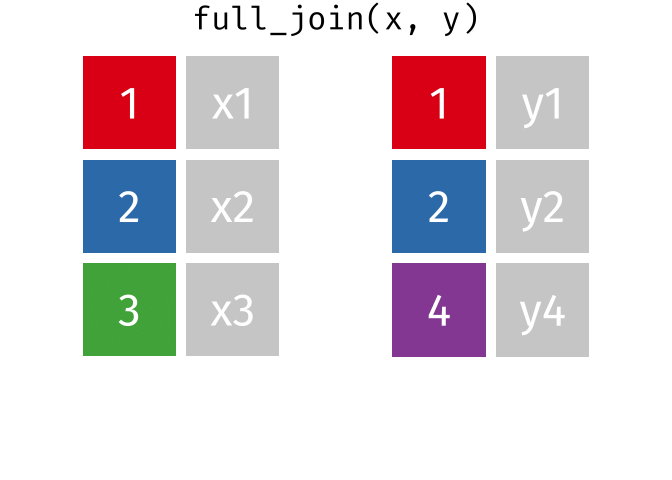
A full_join keeps all rows from both tables, even if a row from the join field isn’t present in one of them. It shows any missing values as NA.
band_members_with_or_without_instruments <-
full_join(band_members, band_instruments, by = "name")
band_members_with_or_without_instrumentsTroubleshooting Joins
Joining when matching columns have different names
Often times, the “join field” in your first table has a different name than that of your second table. For example, you may be trying to join two tables on a common zip code, but the first table calls the column ‘Zip’ and the second table calls it ‘Postal Code’. There’s a special syntax here to make it work:
# Let's change the "name" column to be called "MusicalArtist"
band_instruments_renamed <- band_instruments %>% rename(MusicalArtist = name)
# In our join function, we need to specify that the "name" column in the first table matches up with the "MusicalArtist" column in the second table. We do that by setting our "by" parameter differently:
band_members_and_instruments_2 <-
inner_join(band_members, band_instruments_renamed,
by = c("name" = "MusicalArtist"))
band_members_and_instruments_2You can also join across multiple keys (variables), by giving by a list. It will return only rows that match both variables.
Detecting duplicate values
Duplicate values in your data can cause problems with joins. For example, what if our `band_instruments` dataset listed “John” twice:
# Let's add a new row to band_instruments… say "John" also plays "flute"
band_instruments_with_dup <- band_instruments %>% add_row(name = "John", plays = "flute")
band_instruments_with_dupWhen we try and join our `band_members` table with this new table, we now get 4 rows in the resulting table, even though our first table only had 3 rows… how can that be?
band_members_and_instruments_dup <-
left_join(band_members, band_instruments_with_dup, by = "name")
band_members_and_instruments_dupIn R, if there are multiple matches between the two tables, all combinations of the matches are returned. This GIF illustrates visually how this works:
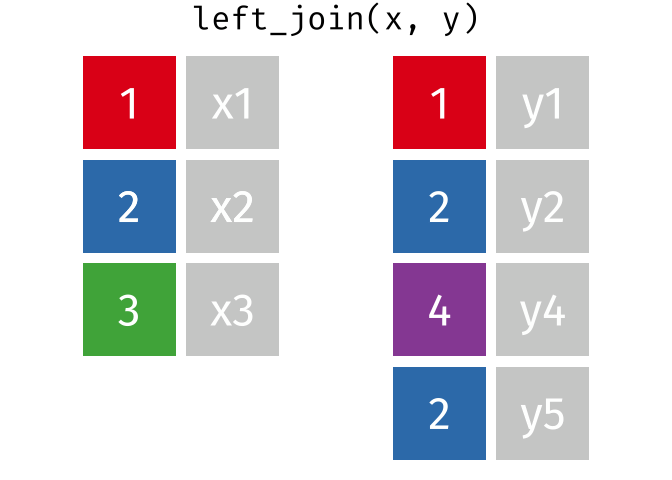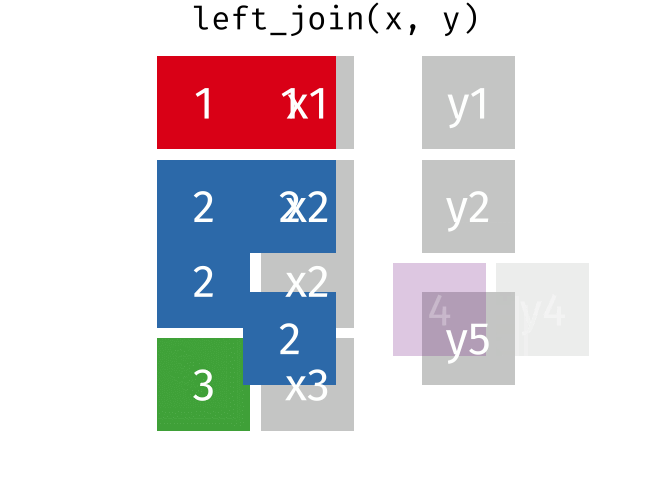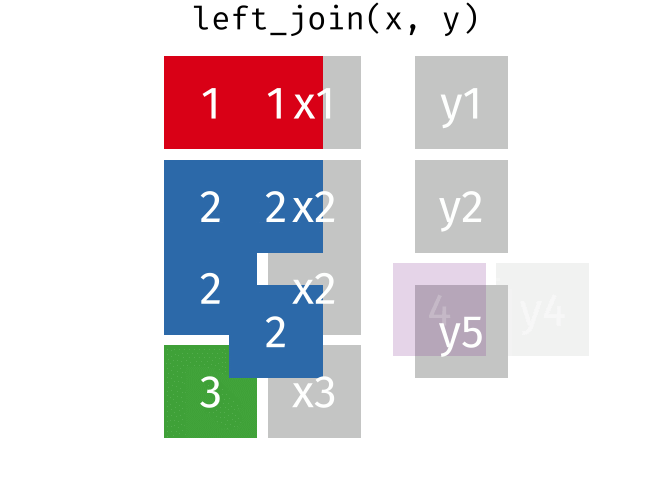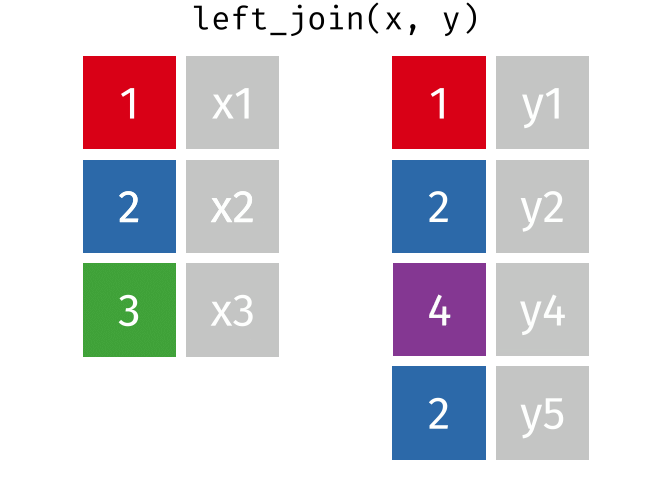
In general, to avoid confusing and unexpected results like this, it’s important to always check for duplicate values in your data, _especially_ in columns that you intend to use as a join field.
Luckily, the `janitor` package has a function for that called `get_dupes`.
library(janitor)# Remember to run `install.packages('janitor')` in your console if you've
band_instruments_with_dup %>% get_dupes(name)Sometimes you may expect duplicate values in a column, and sometimes they may come as a surprise. General Rule: always know what each row represents in your data and what should be unique values.
Sometimes you’ll want to return more rows than you start with, for example if you were matching census divisions to states in order to aggregate the total land area.
regions_divisions #crosswalk between regions and divisions
state_area #this doesn't have the region included! oh no!
regions_states <- left_join(regions_divisions, state_area, by = "division")
regions_states #the join matches state to region and lets us summarize
regions_states %>%
group_by(region) %>%
summarize(region_area = sum(area))